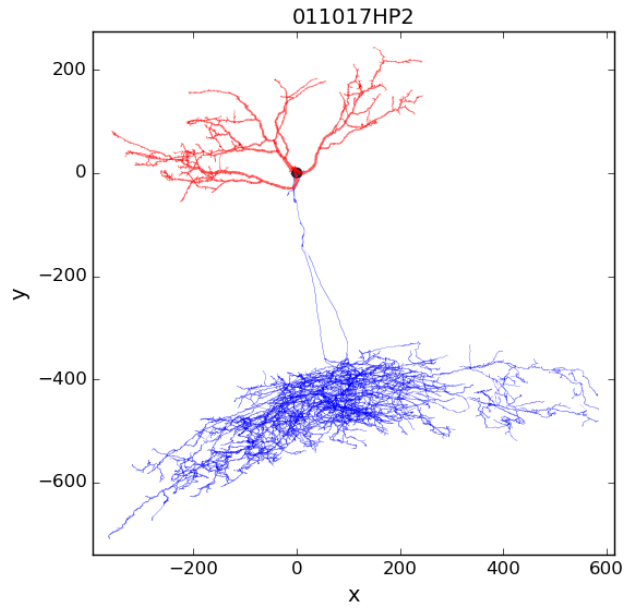
Rebuild an existing single hippocampal cell model
This Use Case allows a user to configure the BluePyOpt to re-run an optimization with his/her own choices for the parameters range. The current version allows to select models obtained in previous optimizations.
Selecting this Use Case, three Jupyter Notebooks are cloned in a private existing/new Collab. The cloned Notebooks are:
Rebuild Single HippoCell - Config
It allows the user to:
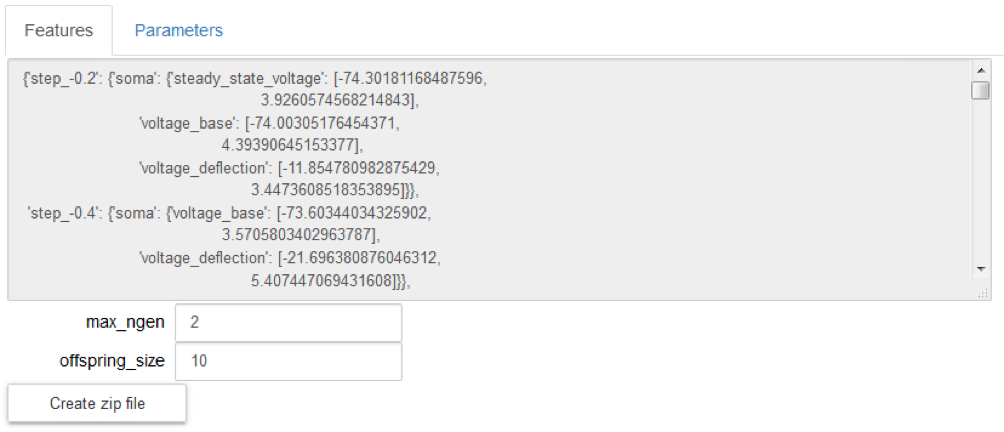
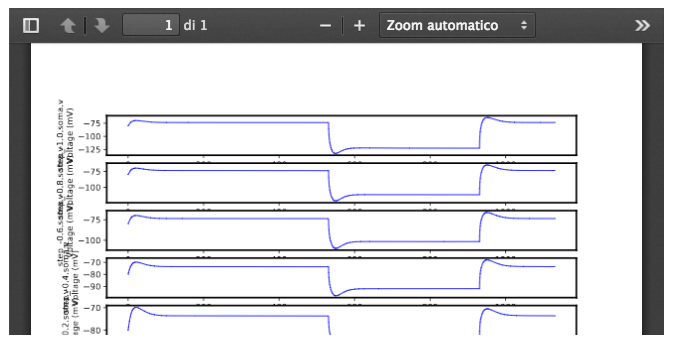
Visualize the electrophysiological features for the chosen model, that will be used as reference by the optimization process (“Features” tab); they cannot be changed:
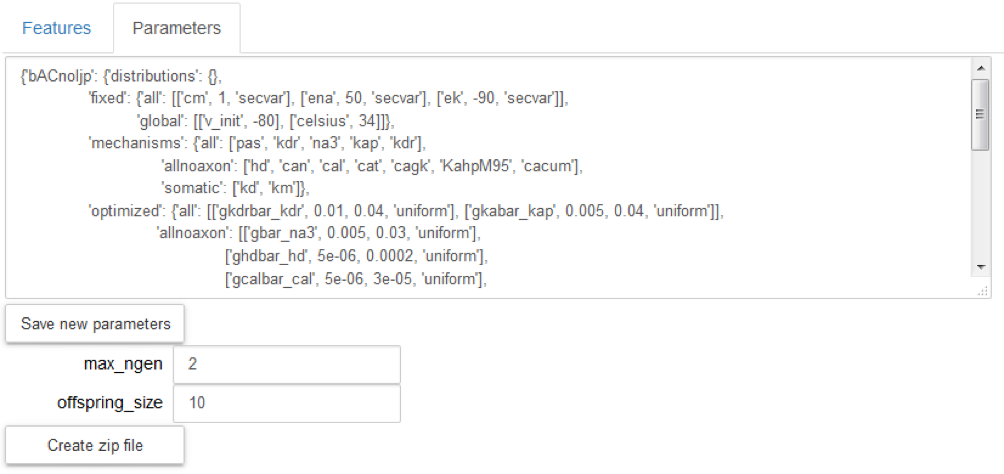
Visualize and change parameters of an existing optimization (“Parameters” tab). Values can be changed and saved by the option “Save new parameters”.
Configure the BluePyOpt optimization algorithm, by defining the offspring size and the max number of generations; they are set by default to 10 and 2 respectively.
Generate a zip file needed to run the new optimization by the option “Create zip file”:
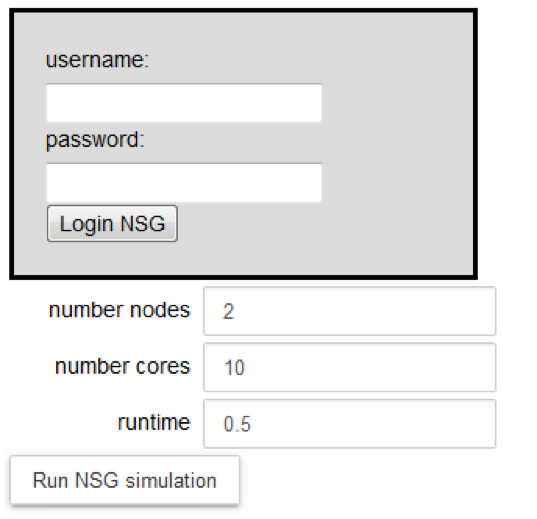
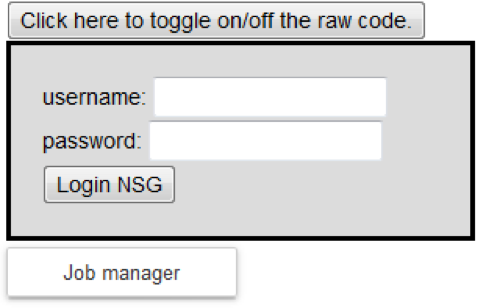
Login to the NSG (Neuroscience Gateway) by inserting a username and password and clicking on the “Login NSG” button:
Configure the parameters of the optimization job: number of nodes, number of cores and runtime. They are set by default to 2, 10 and 0.5 respectively. The maximum number of nodes available per job is 72. If you would require more than 72 nodes, please contact nsghelp@sdsc.edu. The maximum number of cores required per node is 24.
Run the optimization by clicking on the “Run NSG simulation”.
The job has been submitted when you see:
You can check the job status by clicking on the “Check NSG simulation”. The status may be: QUEUE, COMMANDRENDERING, INPUTSTAGING, SUBMITTED, LOAD_RESULTS or COMPLETED; Once the job is COMPLETED, results are saved in the Collab storage, under the BluePyOptOne/resultsNSG/username/foldername (foldername is the name of the old optimization, where date and time are updated with the current date and time).
If you are interested in the code, click on the “Click here to toggle on/off the source code” button:
BuildOwn Single HippoCell - NSG Job Manager
This Jupyter Notebook allows the user to:
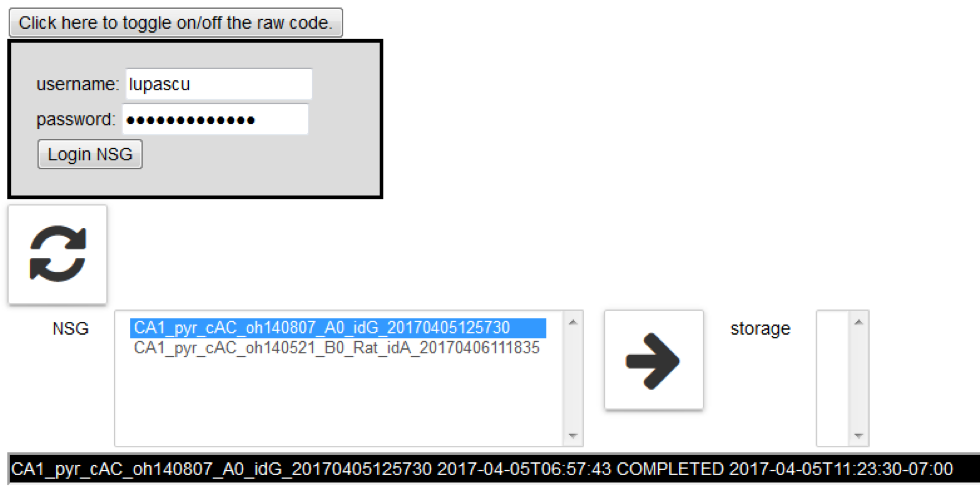
Log in to the Neuroscience Gateway (NSG) and load the Job Manager by clicking on “Job manager”:
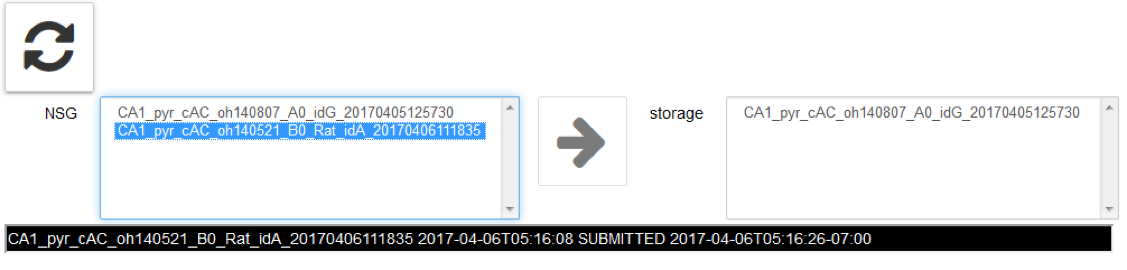
eopy completed jobs from NSG to the Collab storage by clicking on the arrow icon →
Refresh jobs on the NSG by clicking on
 :
:
A job can be stored only if completed:
BuildOwn Single HippoCell - Analysis
This Jupyter Notebook allows the user to:
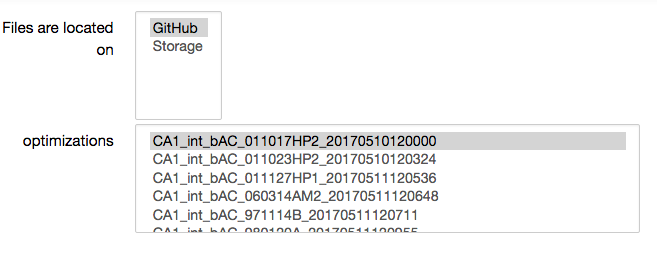
Choose either a previous optimization from the HBP GitHub repository; or choose the result of your own optimization from the Collab storage, and then run an analysis by clicking on “View analysis”
Save the analysis in the storage by clicking on “Save analysis”. The analyses are saved under the BluePyOptAnalysis/username/foldername (foldername is the name of the optimization)
If you are interested in looking at the code, click on “Click here to toggle on/off the source code” button: